Innovation Spotlight: How QIMR Brings Full Traceability to Cancer Research using Semantic Search

Get the latest in your inbox

Get the latest in your inbox
Laboratory Information Management Systems (LIMS) are the nervous center of any research lab. Each LIMS holds patient, clinical, genomic, and trial data, supports research workflows across the lab, and provides the reports required for medical breakthroughs.
Unfortunately, due to increasingly fast-paced research environments, the rigid data models employed by most LIMS can become a bottleneck to innovation in the lab.
To keep pace with the laboratories’ ever-changing data needs and reporting requests, QIMR Berghofer, a leading medical research institute based in Queensland, Australia, transitioned their LIMS from a relational database to a knowledge graph. We spoke with Conrad Leonard, a senior bioinformatician, about how the research at QIMR has improved, how their LIMS has grown 500% in five years, and how the bioinformatics team can now say yes to almost every data request.
Five years ago we were evaluating the technology we wanted to use for our new LIMS. Most of us in the lab have experience with relational database backed LIMS, but we decided to move to graph because it’s very fast research cycle. We need to constantly evolve how we describe data in response to changing hypotheses. Graph suited us well, because in relational databases changing the schema is a long and complex process. But in graph databases we can add new data and new schemas to what we have already built instead of altering existing queries are reports.
Once we settled on graph, Stardog, suited us best because it had the best combination of enterprise features that we needed. Robust authentication, HTTPS, and all the other security bits and bobs that we need. We’re looking into virtual graphs to access our MongoDB instance.
It’s pretty wide ranging. We deal with patient demographic data, patient clinical data, bioinformatics analysis results, and we’re capturing the outputs of internal data pipelines. We’ve just starting to look at incorporating the reporting of our clinical findings as well.
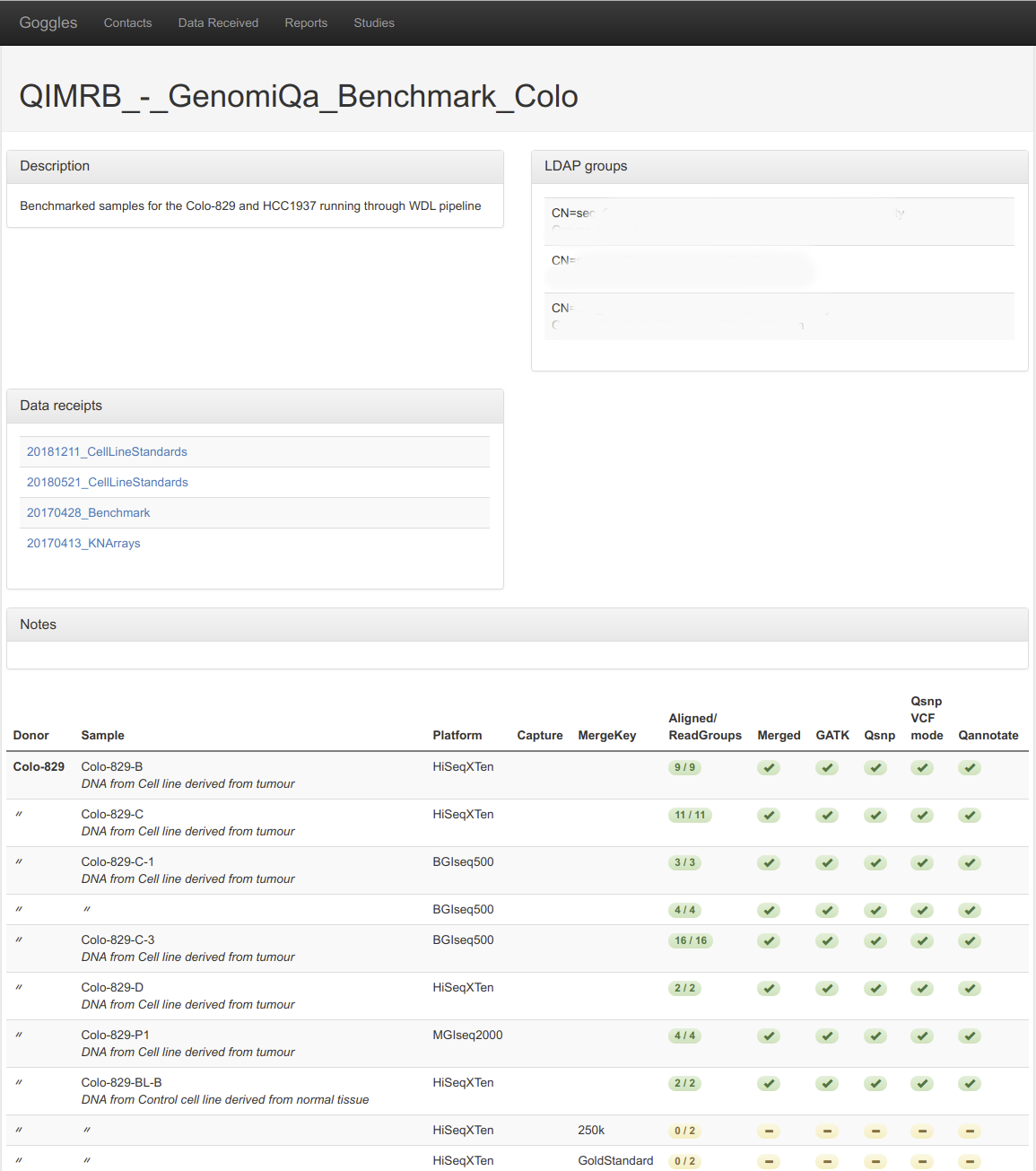
We straddle two research labs, a bioinformatics dry lab and a cancer genomics research lab. Cancer genomics is one of the most data intensive areas of biology. When you sequence somebody’s genome you end up with a couple of multi-hundred gigabyte files, and on top of that you extract the mutations in somebody’s cancer. That can be billions of records per person. From that, you try and extract biologically relevant information to damage pathways or druggable targets. That’s just one patient, we tend to deal with datasets with dozens to hundreds or sometimes thousands of patients. You need a very robust pipeline both in terms of data processing and also capturing metadata, which is what the LIMS does.
Our analysts, programmers, and computational biologists work with the data in Stardog and our management consumes reports generated from Stardog data. For the management, we developed a couple of GUI front ends that present a really big picture: “How many samples in this study have been processed? How much data did we get last month?”
Analysts and biologists are digging into the individual patients and outcomes in the raw data. They have the ability follow the chain of custody and talk about the “why” of the data, which is really important. For example, if we’ve analyzed and retrieved some interesting biology from external data sources and then somebody discovers a problem with a capture kit, we can go back and find out what kit was used for that particular data. Some questions are not the type of questions that would get asked every day, but it’s really important that all the information is captured and is available when we do get them.

It’s been very gratifying to see the keen engagement with this new technology both within the technical team and the wider biology team. Actually, I have been surprised by how quickly the technical people have picked up SPARQL, it’s similar enough to SQL that it didn’t scare anyone away.
The big thing with any LIMS is that you have a single point of truth. You know where your samples are, you know what data you have, what has been done to it. You can drill down to arbitrary levels of precision and know the state of play.
The big improvement to our research is the ability to to drill down very deep. Because of the nature of graph data, you can see the connections between everything. With Stardog you can follow the trail from the sample, to the sequencing library that was made from that sample, to the sequence that was made on that library, to data that came off the sequencing machine, to the next layer of analysis, and the next layer analysis. You can connect all the pieces together very, very easily.
Writing that report or finding that sort of thing out from a relational system can quickly become a nightmare. Once you’ve got more than more than four or five tables involved, the technical requirements of creating all the joins is not fun and certainly not an ad hoc thing. With graph we have the ability to explore the depth of data in a reasonably ad hoc way. That is really powerful.
If analysts or biologists ask us to model some new protocol or change some process, we almost never say “no.” We just say, “yeah, we can do that,” because it’s very easy to extend and to build on what we have.
Before rebuilding our LIMS on Stardog, there were two paths when we got a request that didn’t fit into our data model. Either you shoehorned new data into an existing format that doesn’t quite fit. Or you built another thing and then you have a proliferation nightmare with ten services all dealing with their own specific domain. Both options are ugly.
With Stardog we don’t have that problem. Though, I’m not sure that it is immediately apparent to our biologists. When they ask for something, they expect you to say “yes.”
In five years we’ve increased the size of our data model by a factor of five, and we didn’t start from a very minimal system. We began by replicating what we had running at another lab as our start-up ontology. Every year we add at least a couple of entire protocols, which involve lots of different control vocabulary terms and sometimes entire ontologies.
The lab is looking at incorporating long read sequencing, which is a new technology for looking at structural variants in cancer, and there will be some new protocols and some new datatypes to incorporate into the LIMS.
It’s great not having to fear the unknown. I know there will be something new yet next year; and so, whatever comes, comes.
Is your app powered by Stardog? If you’re building something interesting and would like to be featured in an upcoming Innovation Spotlight, please send me an email!
How to Overcome a Major Enterprise Liability and Unleash Massive Potential
Download for free